“In the field of studying ancient evolution, it’s very hard to falsify or verify. As such people tend to attach unreasonable weight to their beliefs of one possibility or another.” – Julian Gough, father and principal architect of the open-access database SUPERFAMILY
There has been a vigorous and somewhat sharp response to the Charles Kurland and Ajith Harish Tree of Life interview, reminiscent of the email chain circulated by theoretical biologist Stan Salthe following Jerry Fodor’s article in the London Review of Books, “Why Pigs Don’t Have Wings.” I thought it important to share some of the responses to the Kurland and Harish interview, particularly because comments are no longer attached to Huffington Post stories, except on the HP Facebook page.
I received an email via L-form investigator and Leeuwenhoek Medalist Jeff Errington at Newcastle University with comments from his colleague Martin Embley at Newcastle’s Institute for Cell and Molecular Biosciences. Embley has his own Tree of Life.
In my recent interview with Jeff Errington on this page, Errington referred to Embley’s Tree:
“I have a colleague here at Newcastle University named Martin Embley who’s been very prominent in this TOL discussion. Martin and his team have done incisive work that I think supports the idea that there are only two domains and that the eukaryotes are derived from a group within the archaea rather than being an out group and separate from the archaea. The latest textbooks are starting to show the two-domain tree instead of the Woese three-domain tree.”
Martin Embley had this to say in the email Errington forwarded to me:
“As of today a root on the bacterial stem or within bacteria still seems to be the best supported hypothesis and is consistent with other forms of evidence. Kurland and Harish should publish their evidence/analyses in a decent journal like we did and let the community decide how convincing it all is.”
I don’t think Embley was referring to the Kurland & Harish Huffington Post interview, but to Kurland and Harish not publishing their ToL in such scientific journals as Science and Nature, PNAS, etc., as opposed to Biochimie, an Elsevier publication, or to Genome Biology and Evolution, an Oxford University Press journal, where Kurland and Harish have published their deep evolution papers.
Charles Kurland emailed me as follows regarding Embley’s remarks:
“Embley doesn’t seem to realize that with all of his clubby English self assurance that he and his mates have turned Nature into an indecent journal for subjects such as molecular evolution. It is interesting, but he will learn soon that we subjected his phylogenetic model to a statistical test of reliability (Bayesian), and it turns out to be the least probable of the four we tested. . . .”
I once interviewed science and technology historian David F. Noble at length about peer review -- which Noble saw as censorship -- and also covered the now infamous PNAS publishing maneuvers with respect to paper submissions by Lynn Margulis, who at the time (2010) was a member of the National Academy of Sciences.
Kurland said more about the PNAS publishing process:
“Ford [Doolittle] once rejected a manuscript we sent into PNAS because we had not published more papers previously on our data but he had no criticism of what we said in our paper. Peer review is a many splendored thing!”
Speaking of Ford Doolittle— Because Chuck Kurland during our interview said that Doolittle in a 1999 Science magazine article exaggerated the extent to which horizontal gene transfer is a factor in evolution, and then identified Doolitte as a former postdoc of Carl Woese – Doolittle responded with a slur in an email to me, calling Kurland the “Donald Trump of molecular evolution.” (That, by the way, was prior to Trump’s speech to Congress and the Dow surging over 21,000.)
Here are Ford Doolittle’s comments:
“what can I say to that. I was not a postdoc with carl woese and I challenge you to find anything about, or depending on the notion of molecular clocks in my 1999 Science paper. Chuck is the Donald Trump of molecular evolution.”
It’s interesting that Doolittle responded to me regarding Kurland, but did not reply to my request for an interview a couple of years ago when I was putting together my book, The Paradigm Shifters: Overthrowing ‘the Hegemony of the Culture of Darwin’.
Kurland replied directly to Doolittle, copying me on the email:
“Dear Ford;
I am sorry that I identified you as a postdoc with Carl. That mistake arose as my misinterpretation of comments Carl often made to me about your relation to him. Those and your use of the rampant HGT idea in modern organisms, a randomizing principle that he proposed for the progenote, confused me. No dishonor was intended.”
Kurland then excerpted Doolittle’s 1999 Science article:
“After Zuckerkandl and Pauling (4), biologists came to think that the universal tree could be reduced to a tree based on sequences of orthologous genes, any of which (practical considerations aside) could serve as a marker for an entire genome, organism, or species.”
He noted further:
“I think that you will find this a very concise summary of what you would reasonably call a ‘molecular clock.’ It is introduced I suppose to provide contrast to what you say next as a description of rampant HGT:
‘If, however, different genes give different trees, and there is no fair way to suppress this disagreement, then a species (or phylum) can “belong” to many genera (or kingdoms) at the same time: There really can be no universal phylogenetic tree of organisms based on such a reduction to genes.””
Kurland wrapped up:
“I take those two excerpts as the essence of your article, which concludes that there are rampant departures from the constant molecular clock mode of mutations. I say this because I interpret the first situation as requiring constant rates of mutation.
Who is trumping whom?
Cheers/chuck”
Ford Doolittle replied directly to Kurland, copying me on the correspondence:
“Hi Chuck,
Good to hear from you.
Really, of course, I was a sort of spiritual postdoc of Carl’s, and am happy to be so considered. As to Z[uckerkandl] and P[auling], indeed they endorse a clock, but that was not what I had in mind in 1999.
It was the widespread (at that time) assumption, not formally articulated by them as I recall, that all orthologous genes would give the same topology. I also think that we, or at least those of us not actually doing the math, thought at the time that even if different genes evolved at different rates, this would affect branch length but not tree topology.
I find it amazing and somehow satisfying that us old geezers are still engaged in debate.
Regards,
Ford”
SUPERFAMILY father Julian Gough, a bioinformaticist at MRC Laboratory for Molecular Biology, Cambridge whose database Kurland and Harish relied on to build their ToL, agrees that HGT is overblown. Gough wrote to me:
“I agree strongly with some of their [Kurland & Harish] assertions, such as that horizontal gene transfer has been overblown by some segments of the scientific community (because it’s an engaging story and a good excuse for our shortcomings in untangling evolution). . . . I think you capture in your interview something very important, that ‘it’s important that science not become dogma’ and these guys are calling everyone out on some stuff that really has been insufficiently challenged. So I am really pleased that as a journalist you are helping to air the views.”
Since publishing the Kurland & Harish deep evolution conversation, I’ve interviewed Julian Gough. When I asked Gough what he thought about evolution scientists appearing to find the Kurland & Harish model unsettling and being more comfortable with gene sequence analysis, Gough said the following:
“A lot of the objections I see are not so much to the approach but to the conclusions. But often the devil is in the details – i.e., whatever data you use as your starting point for whatever philosophical or technical approach you take.”
The Kurland and Harish ToL challenges the Woese ToL, but there were no defenders of the Woese model, per se, who stepped forward following publication of my interview with Kurland and Harish.
I phoned one high profile scientist who I know is a Woese fan for his reaction to the story, but he didn’t mention the Woese Tree either. He did, however, characterize the Kurland & Harish ToL as “Flat Earth” science, saying it didn’t matter how sophisticated the methods or evidence, the Kurland & Harish conclusions were wrong. He added that I’d made a mistake by doing the interview, and that I shouldn’t have done it.
Bruce Fouke, co-author of the book The Art of Yellowstone Science and a former colleague of Carl Woese at University of Illinois, Urbana-Champaign did email me but said nothing about Woese or about the science. Fouke was an investigator on the five-year $8M NASA Astrobiology Institute-funded project with Woese seeking the principles of the origin and evolution of life. Fouke said simply:
“Thank you Suzan . . . I am following with great interest!!!”
Uppsala University molecular biologist Måns Ehrenberg, an ally of Charles Kurland and Ajith Harish, initially commented:
“Dear Suzan, thanks a lot! Great stuff.”
Ehrenberg later emailed:
“Dear Suzan, there are some remarkable responses among these, e.g., that only Nature published manuscripts or their equivalents can be read and commented upon. In a world where status is the only measure of scientific quality, science is obviously dead. We are not there, yet, but working on in. Take care, Måns”
Mark McMenamin, a paleontologist at Mount Holyoke and former colleague of Lynn Margulis, wrote to me saying he thinks the Kurland and Harish deep evolution scenario “has merit.” Apparently his friend Simon Conway Morris does too, according to Ajith Harish, who spoke with Simon Conway Morris about it at the recent ELSI conference in Tokyo on the emergence of the biosphere.
But McMenamin envisions certain mergers taking place, which he describes as “Margulis writ large” and referred me to the “Pandora’s Pithos” chapter in his book: Dynamic Paleontology for specifics.
However, Kurland and Harish dismiss the Margulis hypothesis of endosymbiosis -- which inspired more comments from Margulis’s allies, among them University of Chicago microbiologist James Shapiro:
“I think Kurland and Harish are mistaken about symbiogenesis and eukaryotic origins. There are too many similarities between eukaryotes and archaea in core information transfer and transcriptional regulation functions for them not to be ancestrally related, and mitochondria are clearly alpha-proteobacteria endosymbionts. Then there are the many photosynthetic eukaryotic groups that originated either in a cyanobacterial or algal endosymbiosis. Those cases cannot be explained in any other way.”
Kurland responded to the Shapiro comment:
“The bathos of the note from Jim Shapiro makes me want to cry. Why do people who have not the slightest notion of phylogeny feel compelled to make judgements about things that they simply do not understand.
Please tell your respondent, Shapiro that there are at least two ways to account for the structural similarities of mitochondrial and bacterial and archaeal proteins.
One is by normal vertical inheritance. The other is by a transfer say from a bacterial genome to the mitochondrial genome (nuclear or organellar). We do phylogeny rather than stare into tea leaves to distinguish these two. The phylogeny is unambiguous so far. There is no evidence of transfer of eukaryote genes from bacteria—NONE. There is strong evidence for vertical inheritance from the common ancestor of more than 90% of the mitochondrial protein determinants.
Of course this has required that we identify the proteome of the last common ancestor. We have done this. But the gene sequence people have not. There is no real evidence that any eukaryote genes had originated in a cyanobacterial genome. Similarity of protein structures does not by itself constitute proof of transfer!!
The 60s was a time when many were discovering drugs and the endosymbiosis model is a fitting product of that period. It is a pipe dream. I have nice feelings about both that period of discovery and that pipe dream. But that does not qualify as a demonstration of the validity of the endosymbiosis model.”
Jim Shapiro was not satisfied with Kurland’s explanation and argued further:
“Neither Kurland nor Harish addresses the photosynthetic lineages, where plastid origins by endosymbiosis are undisputed. The use of emotive terms (as Kurland does) is generally a tip-off that more than a dispassionate analysis of the data is in play.”
Ajith Harish then picked up the thread:
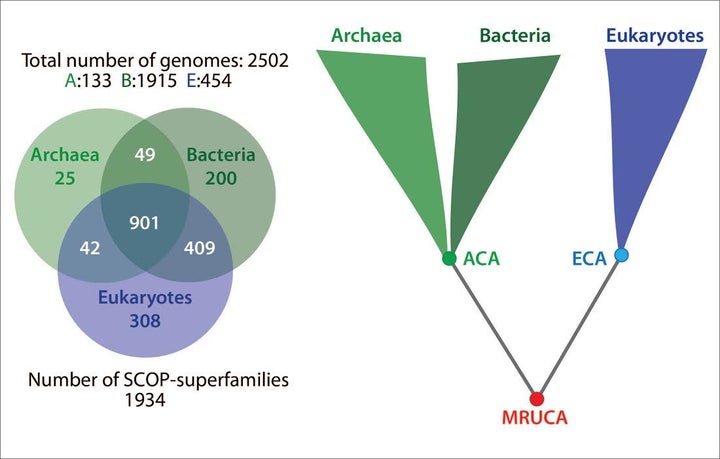
“The data we have so far shows that many photosynthesis-associated superfamilies (SFs) – whether from cyanobacteria or plants or unicellular photosynthetic organisms – can all be traced back to MRUCA [most recent universal common ancestor]. There is a large overlap between mitochondria-associated SFs and plastid-related SFs. But, we need a larger data set to confirm these results. Note that we started out our studies to find the root of the ToL using an objective and rigorous analysis to locate the root despite the common prejudice of where the root “should be.” [W]e sampled a broad range of eukaryotes and bacteria (and archaea). Photosynthetic organisms make up about 25% of the sampling. [W]e want to check with a larger data set. It is on the to-do list.
Phylogenetic analysis is a test of hypothesis, but there have been decades of biased or misguided analyses/interpretations using gene trees where rather than finding an answer, presumptions are simply confirmed. . . . See Keeling’s perspective: “The impact of History on Our Perception of Evolutionary Events: Endosymbiosis and the Origin of Eukaryotic Complexity”. . . .
It has taken decades and armies of gene jockeys to paint the picture we have today. And we [Kurland & Harish] have just started to straighten things out and we are only two of us. It gets very tiring sometimes to answer the same (blind) criticisms, which are raised without reading our papers. Why should anybody read new analyses? Everybody knows what the answer is.
To our knowledge no one has done a rigorous and objective analysis of all the different tree of life proposals (i.e., rootings) and compared them head to head. We’ve done just that.”
Plant physiologist František Baluška at the University of Bonn, who also likes the Margulis endosymbiosis hypothesis, wrote:
“Hi Suzan,
Thank you very much for sending me these links. I am little bit puzzled by the Kurland sentence:
‘. . . the bacteria gets together with an archaea and it makes a eukaryote, the bacteria becomes the mitochondria and the archaea becomes the nucleus cytoplasmic host. That’s the theory. It’s very, very clearly specified that way.’
Especially, of:
‘. . . the archaea becomes the nucleus cytoplasmic host. . .’”
[Note: Kurland’s full sentence was this: “But that’s the essence of the symbiosis hypothesis – the bacteria gets together with an archaea and it makes a eukaryote, the bacteria becomes the mitochondria and the archaea becomes the nucleus cytoplasmic host. That’s the theory. It’s very, very clearly specified that way.]
František Baluška continued:
“This is somehow perplexing – is the archaea leading to both: the nucleus and cytoplasmic host?”
Ajith Harish commented on Baluška’s comments:
“As for František Baluška’s opinions, all I can say is that everything conveyed there is wishful thinking and unsupported speculation. Again, hand-waving arguments are of no help.”
Baluška then said:
“Only time will show how this problem can be solved. I am looking forward to read their new papers when they will be out. From those they have published, I am not convinced.”
Margulis archivist James MacAllister, sent me this message:
“Woese and Fox’s discovery of archaebacterial was certainly fabulously important. When new technology is used, discoveries are made. But Woese’s 3 Domains only make sense when you ignore (as Woese admitted doing) the symbiogenesis that created eukaryotes, eukaryote’s bacteria-derived organelles and the obligate relationship of the holobiont to its microbiomes.”
Virologist Luis Villarreal thought it was “curious” that Kurland and Harish in our interview referred to microbiologist Patrick Forterre at Institut Pasteur but did not speak to “virogenesis.”
But Patrick Forterre in a Frontiers in Microbiology article in July 2015, in which he proposed an updated version of the Woese Tree with several rootings, said he was not indicating viruses in his ToL either, saying further:
“Viruses (capsid encoding organisms) are polyphyletic, therefore their evolution can be neither illustrated by a single tree nor included in the universal tree as additional domains. However, this should not be viewed as neglecting the role of viruses in biological evolution because “the tree of life is infected by viruses from the root to the leaves.”
Forterre, incidentally, regards the ToL as a metaphor, “illustrating the history of life on our planet.”
Villarreal said more:
“Also they [Kurland & Harish] seem unaware of the giant viruses that can infect single cell eukaryotes. I’m attaching two links that might interest you. One on RNA stem-loop cooperation and the other on phage making nucleus-like structures. FYI, the giant DNA viruses also make nucleus-like ‘factories.’”
However, Kurland, Harish et al. did publish an open-access virus article in August 2016 in the Oxford University Press journal Genome Biology and Evolution, titled: “Did Viruses Evolve As a Distinct Supergroup from Common Ancestors of Cells?” in which they write:
“The debate on the ancestry of viruses is still undecided: In particular, it is still unclear whether viruses evolved before their host cells or if they evolved more recently from the host cells. . . .
The recent discovery of the so-called giant viruses . . . revived the reductive evolution hypothesis. . . [E]xamples of common viral components that are analogous to the ribosomal RNA and ribosomal protein genes, which are common to cellular genomes, are not found. This is one compelling reason that phylogenetic tests of the ‘common viral ancestor’ hypotheses seem so far inconclusive.”
Harish has told me he and Kurland hope to include more about viruses in their research as the technology improves.
Brig Klyce at Panspermia.org was excited to see the Kurland & Harish analysis of complexity early:
“The SCOP analysis and plenty of others point to “complexity early.” This does not work at all for Darwinism. But what does work?”
Klyce will be happy to know that as part of “an aspirational training exercise” on directed evolution, Julian Gough’s former students at University of Bristol in the UK are sending extremophile bacteria into space in a CubeSat to “learn how organisms may have evolved ancient pathways to deal with space travel.”
Meanwhile, Binghamton University theoretical biologist Stan Salthe awaits answers for the origin of the gene:
“Thanks Suzan! BTW, what’s the latest on the origin of the nucleic acid gene system?”
